Resources
The Rodrigues lab uses bioinformatic approaches to research where mutations should be made and understand the function of proteins.
ConservFold: Conservation and 3D Structure Generator
Our conservation to 3D Structure/Weblogo pipeline which can all be ran in the browser was developed with collaboration from the Stansfeld lab. It takes the sequence searching and alignment approach of MMSEQ2 and combines this with the entropy calculation of Weblogo to automatically produce images and data on conservation of the protein of interest by sequence across thousands of potential species.
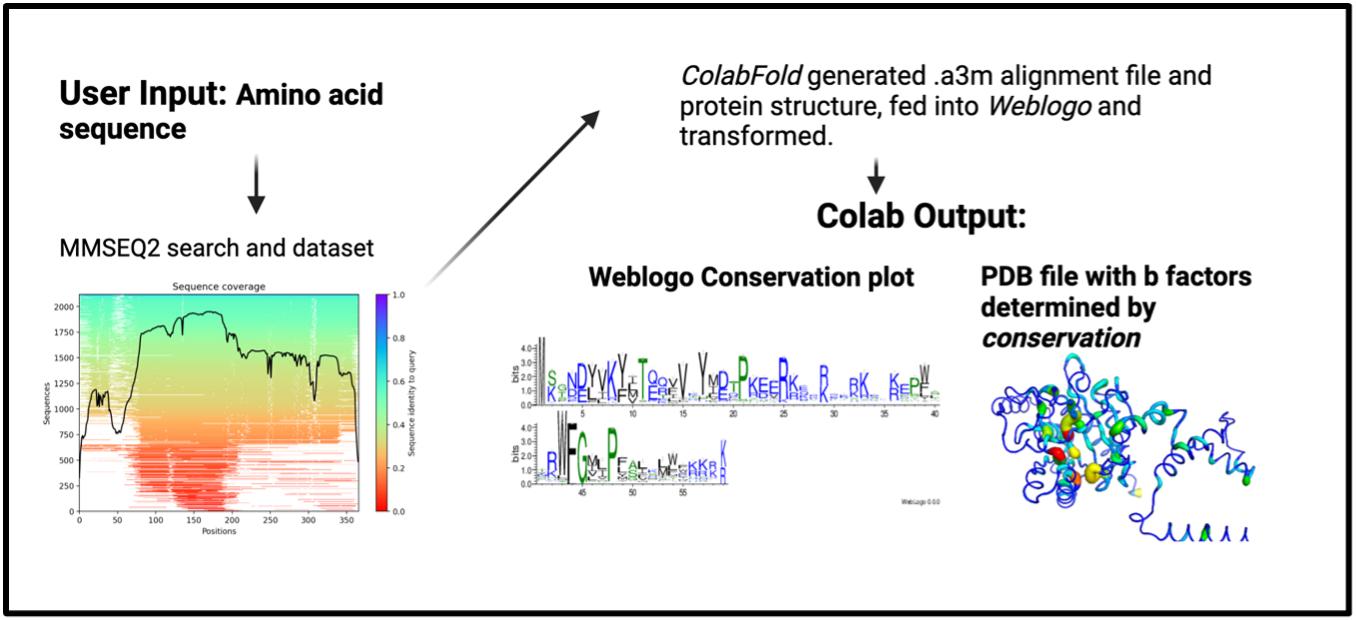
Use our pipeline, and copy the file to your own google colab folder, and run the script with ‘run all’ to run your own predictions by following the associated tutorial. The basis for the sequence conservation is from the previous Weblogo paper. Inspired by the Consurf server we also produce a pdb file and view of the protein with the B factor loaded with conservation. This can also be visualised in protein visualisation programmes such as PyMOL.
Acknowledgments
- We thank the AlphaFold team for developing an excellent model and open sourcing their software.
- Phillip Stansfeld and Chris LB Graham for this notebook combining this with weblogo as well as using weblogo generated conservation to inform the pdb b factor addition.
- KOBIC and Söding Lab for providing the computational resources for the MMseqs2 MSA server. Which this conservation server relies upon.
- Inspired by a colab by Sergey Ovchinnikov (@sokrypton), Milot Mirdita (@milot_mirdita), Martin Steinegger (@thesteinegger),
If you use this software please cite both Weblogo, and ColabFold
If you'd like to cite our simple notebook specifically, which combines weblogo and colabfold elements to automatically assess protein conservation, please use the Github doi here:
